This article is co-authored by Lynn Edens of Snowmass Alpacas and Jason Gill of Cotton Creek Farms.
It was originally printed in Issue 7 of The America Alpaca Journal. You can find the entire June issue on the Snowmass website.
As most of you reading this will know, in early 2021 we began using an alpaca coat color genotyping test offered by the international biotechnology company Neogen Corporation to learn our animals’ base coat color genotypes. This test is based on research by Dr. Kylie Munyard and specifies alpacas’ genotypes for relevant sections of the two most important genes affecting alpaca base coat color. It also reveals whether the animal carries a mutation that produces the tuxedo grey pattern.
Knowing the base coat color genotypes of our animals has already made a significant difference in our breeding decisions and has also helped us more accurately select the animals in our herd that are best suited for our customers’ breeding programs. However, the results of our first rounds of testing, as well as those of other farms who have joined us in the research effort, also made it clear that we still had much to learn, especially with respect to the genotypes that produce animals with a black base coat color.
We knew from reading Dr. Munyard’s research that she had identified three different alleles — that is, alternate forms — of one gene, known as the agouti or ASIP gene, that when paired produced a black animal at least some of the time. These alleles were collectively described with a small “a” in the reports we received from Neogen. If an animal’s ASIP genotype included one small “a,” we knew it covered black. If an animal carried two small a’s, it had a black ASIP genotype and, assuming it was not fully dilute, could be phenotypically black.
However, only a bit more than half of the not fully dilute animals in our database with “aa” test results actually had true black, bay black, or silver grey coats (silver greys being black animals with a separate white pattern.) Almost as many were brown or rose grey. This suggested the possibility that the three individual ASIP “black” alleles might differ with respect to their impact on phenotypic color.
We asked Neogen if they could go beyond the summary results previously presented and give us the detailed genotyping results for the animals they had tested for us so we could study this further. They could and did. Other breeders who had genotyped their herds with Neogen also requested their detailed results and contributed them to our research database. From this collective effort, we learned that the three individual black alleles do not have identical impacts on phenotypic color. Two of them appear significantly more likely to produce a black animal than the third.
Understanding how all three of these alleles work, in both homozygous and heterozygous pairs, greatly improves our ability to predict color outcomes from individual matings. We now know how to more consistently produce black alpacas, and how to focus on true black and minimize the production of bay blacks where that is desired. We also have a better idea of how to best choose animals that are not black but cover it for a black breeding program.
We discuss these results in detail here so you can use them to inform your own breeding efforts.
The ASIP Gene’s Black Alleles
The acronym “ASIP” stands for agouti-signaling protein. The ASIP gene encodes the instructions for producing the agouti-signaling protein, which controls the amount of pheomelanin (yellow) versus eumelanin (black) pigment produced by the melanocytes. When the agouti-signaling protein produced is normal in form, both pheomelanin and eumelanin are produced by the melanocytes in proportions which, in the absence of dilution, results in an animal with a dark fawn or brown base coat color. However, when a mutation results in the production of a protein that is altered in a way that makes it less functional, less or no pheomelanin and more eumelanin is produced by the melanocytes, resulting in an animal with a darker or black coat color.
Something else happens, too: When the agouti-signaling protein is not active in the melanocyte, a hormone called the melanocyte-stimulating hormone plays a greater role in pigment production. This hormone does exactly what its name suggests, with the result that the melanocytes produce more pigment than they otherwise would. Animals with “aa” genotypes thus not only have more of the black eumelanin pigment, but a greater absolute amount of pigment in their fiber.
The three ASIP alleles that have been associated with black coat color in alpacas are all of this “loss of function” type and result in the melanocytes producing less pheomelanin than they otherwise would. Each of these alleles involves a change to a particular section of the ASIP gene known as Exon 4. Exons are segments of DNA which contain the instructions for producing a protein. Exon 4 of the alpaca ASIP gene is 177 base pairs long. You don’t have to know what this means to find it interesting context for what follows. You may also find the picture shown in Figure 1 helpful for understanding the discussion that follows — we did when it was presented to us.
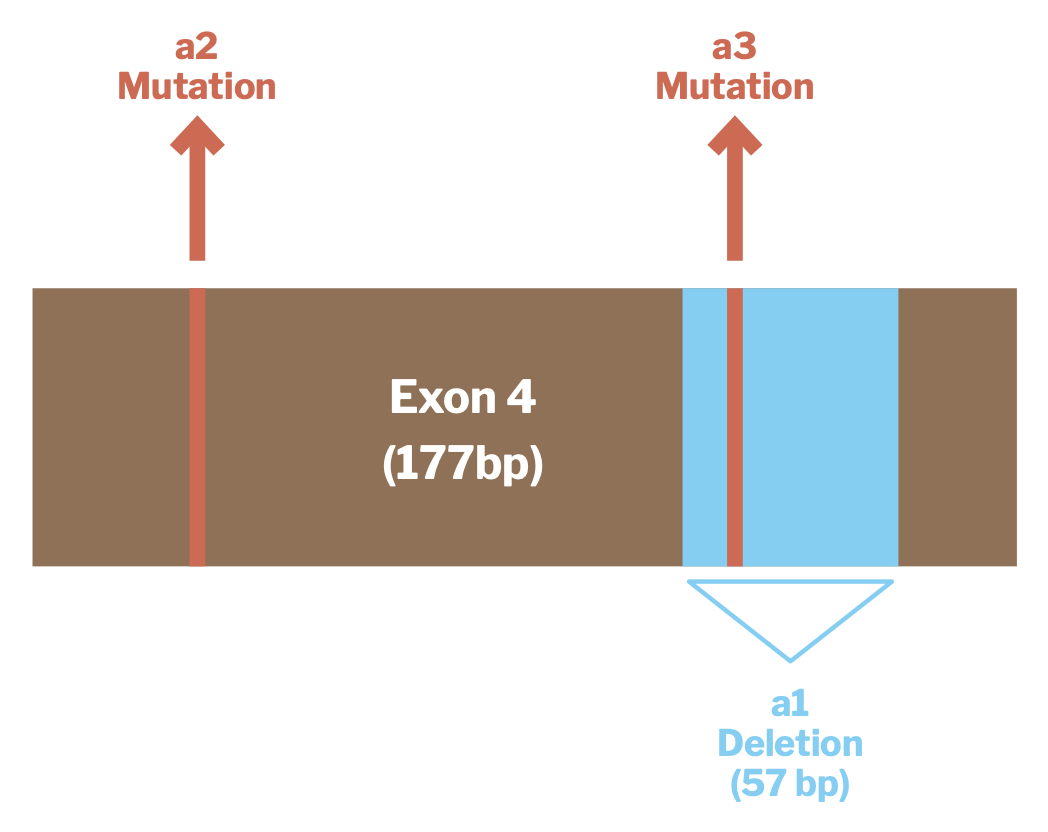
Source: Feeley et al, Three novel mutations in ASIP associated with black fibreVi in alpacas (Vicugna pacos). J Agric Sci 149: 529-538
The genotypic alternation that produces the allele that, following Dr. Munyard’s terminology, we describe as “a1” involves a deletion of 57 of Exon 4’s 177 base pairs. As you can imagine, that represents a substantial disruption to the coding for the agouti-signaling protein. The “a2” and “a3” alleles both involve mutations at individual base pair locations in Exon 4. Both occur at locations where mutations are known to result in some loss of function of the agouti-signaling protein. Interestingly, the location of the “a3” mutation is within the region that is deleted and thus missing from the “a1” genotype.
All phenotypically black animals have two of the black alleles described above: Two of one allele type, or one each of two different black alleles.
Allele Combinations And Their Phenotypic Results
To discover potential differences in the phenotypic results produced by different black allele pairings, we combined the detailed genotyping results of animals at four farms to produce a dataset of 181 animals that had “aa” ASIP base coat
color genotypes and carried either zero or one Mc1R gene dilution mutations. That is the animal’s base coat color genotypes were summarized as either EE aa or Ee aa, both of which can result in a black animal.
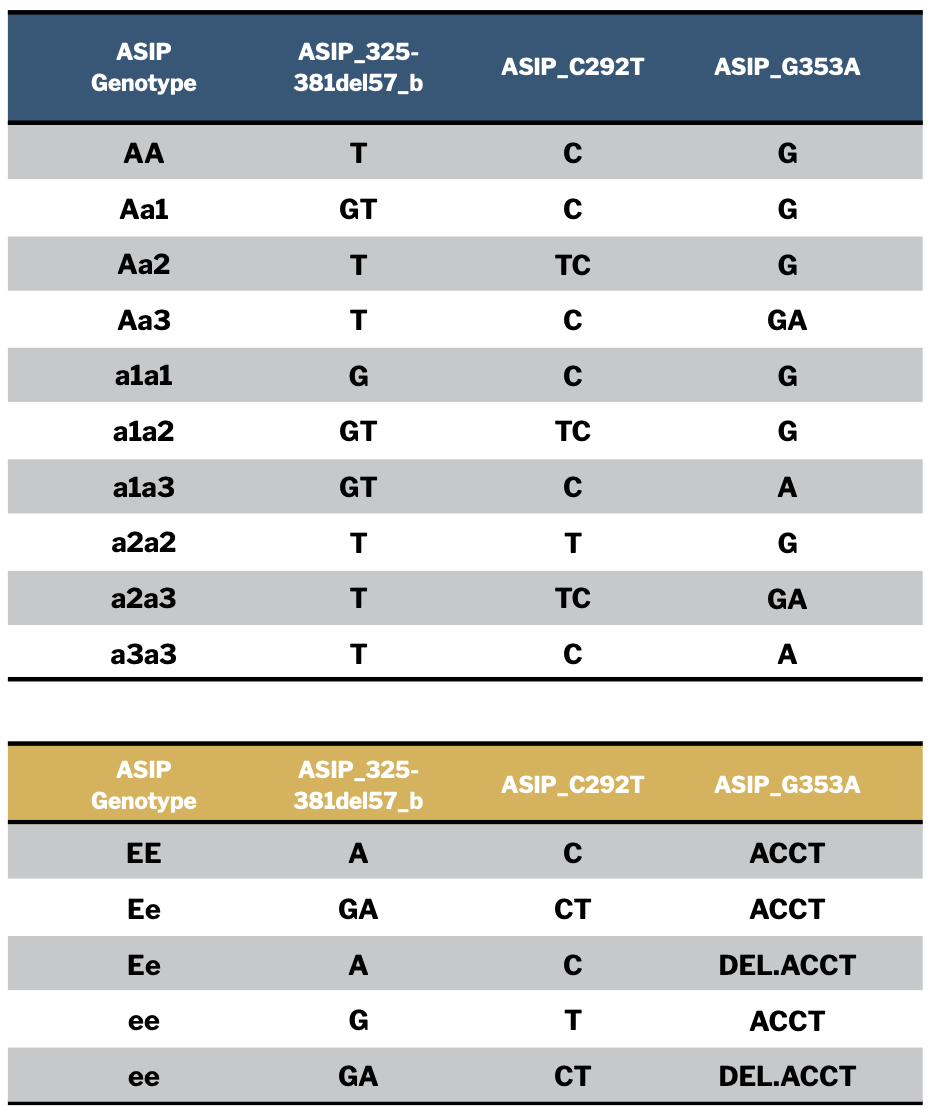
However, there are actually 12 possible combinations of the three “a” alleles and the two dilution genotypes EE and Ee. They appear in the table in Figure 3 along with the specific allele combinations with which they are associated. You can use this table to interpret your own detailed Neogen report results if you choose to color genotype your animals.
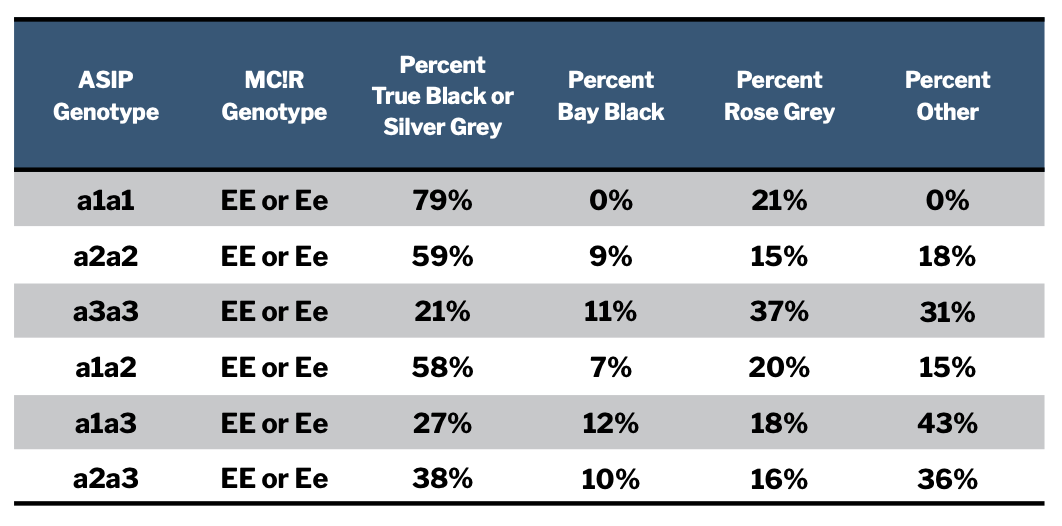
The table in Figure 2 summarizes coat colors of animals with each of these different “aa” genotypes. We chose the way we tabulated the results with some care. True black and silver grey animals are counted together because both tuxedo and modern silver grey animals are the result of white patterns expressed within a black base coat color. We counted bay blacks separately from true blacks, even though they show together, to see if we could identify specific genotypes that were more likely to produce one versus another. We counted rose greys separately from browns because their genotyping results were not what we expected and we wanted to consider them more closely, which we do later in this discussion. All animals with EEaa or Ee aa genotypes that were not one of these colors we included in the “other” category. The great majority of these were brown.
It can be very tempting to draw some rapid conclusions looking at a table like this, but it is important to first assess the statistical significance of the associations we think we see. We want to know how likely it is that we are observing a real relationship between genotypes and color outcomes as opposed to a pattern produced by chance. We will explain our degree of confidence in the patterns we see as we discuss them.
Based on the genotyping results for the 181 animals considered here we are highly confident that an “aa” animal which has one or two of the “a3” ASIP alleles is less likely to be registered as true black or silver grey (black base coat) than one that does not have an a3 allele. The odds are less than 0.1% that this pattern we see in the data is due to chance. Black ASIP genotypes that include one or two a3 alleles are also more likely to produce bay black animals than are genotypes which lack an a3 allele. More broadly, over two-thirds of the animals with an “aa” genotype in our sample group that do not have black fleeces carry at least one a3 allele.
We don’t know why this is the case but suspect that the a3 mutation either does not reduce the proportion of pheomelanin produced to the same degree (leading to a warmer, browner coloration), does not allow the melanocyte-stimulating hormone to assume as much of a role in pigment product as do the other black alleles, or both.
There is also a pattern in the data that suggests that the a1 ASIP allele may be more likely to result in a true black base coat color than the a2 allele. However, we don’t yet have enough individual observations to be confident that this pattern is not the result of chance. This means that for now, we cannot reliably differentiate between the likely phenotypic coat colors produced by a1 and a2 alleles. We assume they have a similar likelihood of producing black animals. This conclusion may or may not change as we collect more genotyping results.
Finally, we looked at whether carrying a single dilution mutation — that is, having an “Ee” MC1R genotype — reduced the odds of an animal having a true black base coat color. Again, this is a pattern we think we see in the data. Based on the distribution of results and the number of observations we have now, there is about a 90% chance that a single dilution allele does have this effect. Those odds do not rise to the level of confidence typically sought by scientific researchers, but this nonetheless likely effect of dilution is worth keeping in the back of your mind while we collect more data.
Even more important to understand, however, is that even if carrying a single “e” MC1R mutation does not result in different phenotypic coat color for an “aa” animal, it makes an enormous difference in the colors of the crias that animal can produce. An animal with an “EE aa” genotype will produce offspring of fawn or darker base coat color regardless of the color genotype of the animal to which it is bred. By contrast, an animal with an “Ee aa” genotype can produce crias in the whole spectrum of base coat colors, including white and light, depending on the 8 color genotype conveyed by the other animal in the pairing.
Using This Genotype Information
Based on what we have learned so far, we believe that breeders focused on producing true black and silver grey animals should have a preference for breeding animals that carry the a1 and a2 ASIP mutations relative to those that carry the a3 mutation, all else constant. That last phrase is very important. Animals carrying one or two a3 mutations still produce offspring that are registered true black and silver grey, albeit at a lower frequency than those animals with “aa” genotypes that do not include an a3 mutation. We would not exclude them from true black breeding programs, but rather make sure they provide additional incremental advantages to compensate for their less frequent production of the desired true black base coat color. Superior genetics for other fleece traits, genetic diversity value, and potential branding and marketing advantages are among the potential reasons to include these animals in a true black breeding effort.
We want to stress, too, that breeders will increase the number of black animals they produce from other black animals if they maintain as many animals as possible that do not carry a dilution mutation. An “EE aa” animal bred to another “EE aa” animal will never produce a light fawn, for instance, while pairing two phenotypically black, “Ee aa” animals will produce a light fawn 25% of the time. Again, though, animals with black ASIP genotypes and one or even two dilution alleles may bring other important attributes to the program that make the reduced odds of true black offspring worth the trade-off. For instance, we are excited to breed an exceptional young light fawn male with an ee a1a3 genotype to black females in the coming season: His fleece is incredibly dense, long and uniform, and he will bring important genetic diversity to the colored breeding effort at Snowmass.
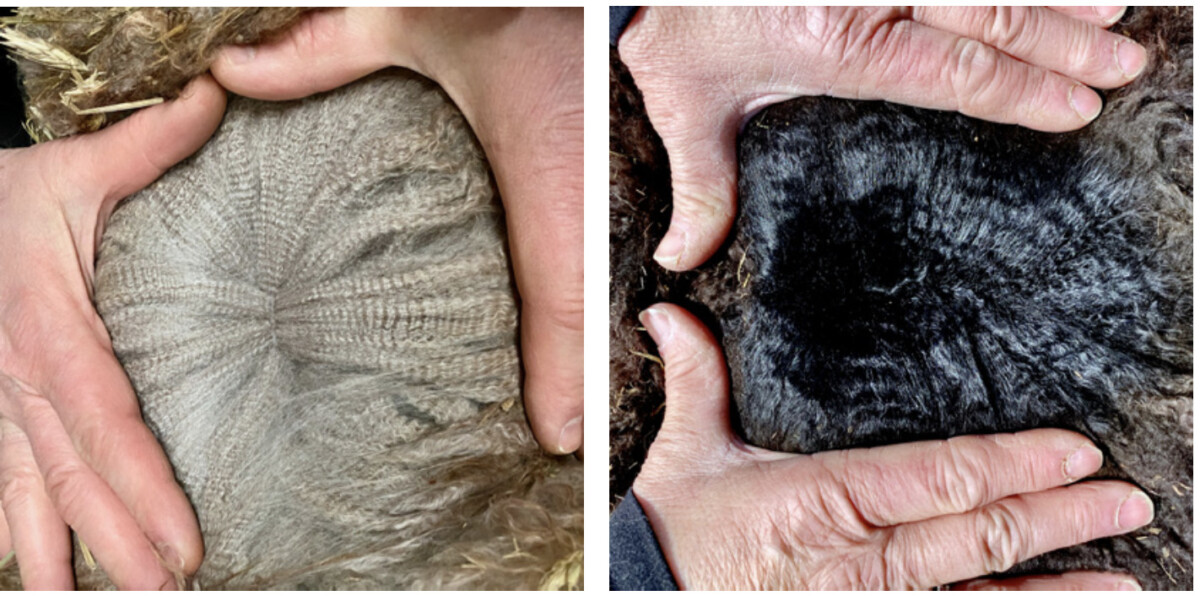
Right image: The fleece of an animal with an EE a1a1 MC1R/ASIP genotype.
The Rose Grey Base Coat Color Genotype
As we mentioned earlier, the base coat color genotypes for the 51 rose greys for which we had detailed genotyping results surprised us. Of those 51, close to two-thirds had “aa” genotypes. We presumed that we would see a much higher proportion of rose greys — which we perceive as brown animals with a white pattern — that had ASIP genotypes of Aa or AA, because many brown animals have these ASIP genotypes. Instead, an “aa” ASIP genotype was most common, and of the rose animals in our sample only four that had been registered as rose grey did not cover black. When we went back to check the phenotypes of those four, we discovered they were actually all fawns with some white contamination in their blankets. They would not be considered rose grey for show purposes, or by most breeders.
When we compared the base coat color genotyping results of rose grey animals to those of dark brown animals, we saw similarities. Almost half of the dark brown animals we have tested to date have also “aa” ASIP genotypes, and 90% of them carry at least one “a” allele and thus cover black. By contrast, only 18% of the nearly 100 medium brown animals we have color genotyped have black ASIP genotypes. Seventy-five percent of the medium browns we have tested carry at least one “a” allele.
It is interesting that rose grey animals have base coat color genotypes that in their distribution most closely match those of phenotypically dark brown animals, and that the animals that would be described by broad consensus as rose grey in our sample all carry at least one black allele. We would have imagined that an animal with an EE AA color genotype, which typically produces an animal with a base coat color of dark fawn to medium brown, along with the white pattern produced by classic grey or roan mutations would also appear rose grey to the typical observer.
However, we have not uncovered any such animals to date. This makes us wonder if the presence of at least one black ASIP allele is required for animals to reveal a pattern that we would describe as classic or modern rose grey based on AOA’s current color classification system. Many rose greys have fleeces that combine black, brown and white fibers. The top fleece image shown below, which is from an animal that would show as a light rose grey, is a good example of this. This may be a clue.
The Blue Black Genotype
One of our personal hopes for our study of black genotypes is that the results will reveal the genotype(s) responsible for producing animals with fleeces we would describe as blue black. There is not yet agreement or even much discussion about what a blue black alpaca looks like compared to true black. While we hope to eventually quantify degrees of blackness with results from a spectrophotometer (the international testing laboratory SGS, which many of you use for histogram analyses, can provide spectrophotometer analysis as well) our sense of the blue black phenotype is as follows: 1) the animal appears an especially deep, cool black even compared to other true black animals, and 2) its fleece looks
black even when compared to dyed black fabrics (some true black fleeces often look brown when compared to commercially dyed fabric.)
These animals are not common in the United States. And they share other fleece attributes besides their exceptional black coloration, because the greatly increased amount of total pigment in the hair shafts of the fibers of a blue black fleece reduces the amount of curvature that can be expressed. These fleeces are typically silky in style with a bold, low-frequency crimp, like that shown by the cria fleece. Their lack of curvature makes them comparatively bright as well (and, we have learned, difficult to photograph).
We asked the breeders who submitted their animals’ detailed genotype information to help build our database to indicate which of their animals they felt were blue black in color. Of the more than 1000 animals for which we have detailed ASIP genotypes, only 11 were identified as blue black by their owners. (Their individual standards for what constitutes a blue black likely vary to some degree.) None of those animals carried an a3 allele, and we suspect this may be an important aspect of the blue black genotype. Eight of the 11 carried at least one a1 allele, and four were homozygous a1a1. And 9 of the 11 did not carry any MC1R dilution mutations.
Digging in a bit more, of the four animals in their own herds that these authors identified as blue black using the general criteria described above, all were of the EE a1a1 genotype, including the cria whose fleece is shown in the picture.
Thanks to the willingness of other breeders to combine their genotyping results with our own, we have collectively made great strides in understanding which ASIP and MC1R genotypes are most likely to produce alpacas with black base coats. But even with our improved understanding of the likely impact of the a3 mutation on coat colors, we still can’t explain why 4% of the animals that have “EE aa” genotype and lack an a3 allele are not true black or silver grey but are instead rose grey or brown.
Other mutations, in ASIP or perhaps other genes, may provide an answer. We are optimistic that those answers will be forthcoming, too, because U.S. breeders’ interest in and quick adoption of color genotyping has helped stimulate more research on this topic. We will continue to share what we learn — and what we are taught — about alpaca coat color genotypes in the months ahead.
Thank you to all the farms who have participated in the process. Your willingness to share your genotyping data has helped the alpaca industry grow in knowledge and improve our collective breeding efforts. Participating farms include Black Barn Alpacas of Texas, Cinco C’s Alpacas of Pennsylvania, Claddaugh Farm Alpacas of New York, Cotton Creek Farms of Michigan, Fun In The Country Alpacas of Michigan, Holdfast Hilty Family Farm of Ohio, Paragon Alpacas of Maryland, Seven Acres Alpaca Farm of New York, and Snowmass Alpacas of New York.






Excellent article. We’re using the observations in our breeding program.